Abstract
Facultative asexual reproduction is a trait commonly found in invasive species. With a combination of sexual and asexual reproductive modes, such species may adapt to new environments via sexual recombination during range expansion, while at the same time having the benefits of asexuality such as the maintenance of fitness effects that depend upon heterozygosity. In the Western United States, native species of Rubus (Rosaceae) reproduce sexually whereas exotic naturalized Rubus species reproduce by pseudogamous apomixis. We hypothesized that new asexual lineages of Rubus could arise from hybridization in this range. To detect hybridization between native and exotic Rubus, we genotyped 579 individuals collected across California, Oregon and Washington with eight nuclear microsatellites and two chloroplast markers. Principal Coordinate Analysis and Bayesian clustering revealed a limited amount of hybridization of the native R. ursinus with the exotic R. armeniacus and R. pensilvanicus, as well as cultivated varieties. Genetic distances between these hybrids and their offspring indicated that both R. ursinus × R. armeniacus and R. ursinus × R. pensilvanicus produced a mix of apomictic and sexual seeds, with sexual seeds being more viable. Although neither of these hybrid types is currently considered invasive, they model the early stages of evolution of new invasive lineages, given the potential for fixed heterosis and the generation of novel genotypes. The hybrids also retain the ability to increase their fitness via sexual recombination and natural selection. Mixed reproductive systems such as those described here may be an important step in the evolution of asexual invasive species.
Similar content being viewed by others
Introduction
Asexual reproduction is commonly seen in invasive species, sometimes with an entire invasion consisting of a small number of clones or even a single clone (for example, Poulin et al., 2005). This is unexpected given that genetic diversity and sexual recombination are generally assumed to be required for natural selection and adaptation to novel environments (Lee, 2002; Barrett et al., 2008). However, it was recognized early in the field of invasion biology that an asexual organism with broad ecological tolerance can be a successful invader, particularly if asexual reproduction maintains heterosis or other fitness effects that depend on heterozygosity (Baker, 1965). Modern molecular studies have confirmed the existence of single invasive clones with broad environmental tolerance and phenotypic plasticity (for example, Geng et al., 2007). Asexual reproduction assures that even a single founding individual can establish a new population and prevents the loss of alleles and the appearance of deleterious recessive phenotypes that come with inbreeding (Barrett et al., 2008). It has been suggested that asexual reproduction with occasional outcrossing is an ideal scenario for the production and maintenance of highly aggressive genotypes (Novak and Mack, 2005), which would be expected if the high fitness of a genotype depends on particular allele combinations that would be lost with sexual recombination.
A case in which asexual reproduction can be particularly beneficial to an exotic species is that of first-generation hybrids. These hybrids may have increased fitness due to dominance, overdominance and epistasis effects, creating a situation in which sexual reproduction is disadvantageous (Springer and Stupar, 2007). Hybridization has led to the evolution of many new invasive taxa, many of which are successful due to putative fixed heterosis as a result of asexual reproduction or allopolyploidy (Schierenbeck and Ellstrand, 2009). Hybridization between a native species and a naturalized exotic species can produce fitness benefits in addition to heterosis, as the native species may have adaptations specific to its environment, and the exotic species may have an overall high fitness and broad environmental tolerance that allows it to persist in the novel habitat.
Plants are especially capable of asexual reproduction, making them likely candidates to maintain the fitness advantages of hybrids through fixed heterosis. In addition to vegetative propagation, plants may reproduce asexually through apomixis (syn. agamospermy, asexual seed production). Apomixis appears to be frequently associated with polyploidization, or can be inherited in a dominant manner when sexual and apomictic plants are crossed (Ozias-Akins and van Dik, 2007). This suggests three mechanisms by which apomixis can follow interspecific hybridization: allopolyploidy following a cross between two sexual species, dominance when a facultative or pseudogamous apomictic species crosses with a sexual species, or a rare sexual event between two facultative apomictic species.
The genus Rubus (blackberries and raspberries) includes many invasive species worldwide (see the USDA PLANTS database (United States Department of Agriculture, 2011) for many examples in the United States alone) that are taxonomically well-distributed across the genus. There is one documented example of hybridization followed by apomixis in the history of an invasive Rubus species: after introduction to Madagascar from Southeast Asia, Rubus alceifolius hybridized with the native R. roridus to produce apomictic lineages that then invaded other Indian Ocean islands (Amsellem et al., 2001). The most widespread group of invasive Rubus, the R. fruticosus aggregate of species, also reproduces primarily by asexual means. This species complex is native to Europe, and consists of a few sexual diploid species and many tetraploid clones that reproduce by facultative pseudogamous apomixis and occasionally hybridize with each other (Weber, 1996). Evans and Weber (2003) identified only two invasive clones of R. fruticosus agg. across Australia, New Zealand and the United States, suggesting that preadaptation to a broad range of environments may compensate for very low genetic diversity in asexual lineages of invasive Rubus.
Both native and introduced Rubus can be found in the Western United States. The most common native species, R. ursinus, is generally diecious and has hexaploid, octaploid, dodecaploid forms (Brown, 1943) and has been hybridized with other species to produce several popular cultivated varieties (McGregor, 1998). Diploid native species include R. leucodermis and R. spectabilis in the subgenus Idaeobatus and R. parviflorus in the subgenus Anoplobatus (Thompson, 1995). All of these native species reproduce sexually (Crane, 1940). Most exotic naturalized Rubus in this range belongs to the R. fruticosus agg. (subgenus Rubus). Within this species aggregate, R. armeniacus (syn. R. discolor and R. procerus) is the most widespread invasive type in the Western United States, and several other microspecies are naturalized but not nearly as widespread (United States Department of Agriculture, 2011). Outside of R. fruticosus agg., R. pensilvanicus (subgenus Rubus) is naturalized in the Western United States after introduction from the Eastern United States (Ertter, 1993). Both of these exotic types are tetraploid and reproduce primarily by pseudogamous apomixis, with the exception of one R. fruticosus agg. microspecies, R. ulmifolius, which produces seeds sexually (Nybom and Schaal, 1990; Thompson, 1995; Weber, 1996). Rubus is also grown as a fruit crop in this geographic region, with cultivars derived from R. ursinus, R. fruticosus agg. and other species of the subgenera Rubus and Idaeobatus, including a number of hybrids between these subgenera (McGregor, 1998). Spontaneous hybridization between native and exotic Rubus in the Western United States was documented in the mid-twentieth century (Mallah, 1954) and continues to be observed by modern botanists (Lawrence Alice, personal communication).
In this study, we test the hypothesis that ongoing hybridization between native, exotic and cultivated Rubus taxa in California, Oregon and Washington is producing novel apomictic lineages. We first determine the amount of spontaneous hybridization and introgression within natural populations using nuclear microsatellites and PCR-based chloroplast markers. We then determine the reproductive mode of the hybrids by examining microsatellite genotypes of offspring of three spontaneous hybrid clones. More broadly, we characterize a system that reflects current models of the evolution of invasiveness. In this system, hybridization allows retention of asexual reproduction that contributes to the introduction success of an exotic parent, while at the same time incorporating native genetic material that is locally adapted and allowing further adaptation by sexual recombination. The system of hybridizing Rubus in the Western United States has the potential to provide insight into how novel invasive hybrids may evolve.
Materials and methods
Materials for genotyping
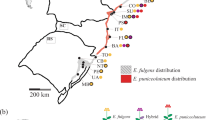
To test for hybridization and introgression in natural populations, leaf tissue was collected from 552 Rubus plants at 35 sites across California, Oregon and Washington, USA (Figure 1, Supplementary Table 1). Samples were collected along one-dimensional transects, usually following a stream or trail, because blackberry thickets can be impenetrable and are often restricted to the edge of a body of water. As the growth habit of Rubus makes it difficult to determine where one individual ends and another begins, randomly generated numbers between 5 and 50 were used to determine the number of paces between samples of the same species. Pressed voucher specimens were deposited at the University of California-Davis Center for Plant Diversity. Additionally, 23 samples from cultivated varieties were collected from two commercial farms and a student experimental farm in California (Supplementary Table 1) and four cultivated specimens were obtained from Brooklyn Botanic Garden in New York. Each leaf sample was dried and stored in silica gel (Chase and Hills, 1991) until DNA extraction.
To determine the reproductive mode of hybrids, seeds were obtained from hybrids and parent species. Cuttings of two putative R. ursinus × R. armeniacus clones from collection site CSP (Supplementary Table 1), R. pensilvanicus from BWP and one putative R. ursinus × R. pensilvanicus clone from BWP were treated with Hormex Rooting Hormone #3 (Brooker Corporation, Chatsworth, CA, USA), placed in a 1:1:1 perlite:vermiculite:soil mix on a heated mist bench, then after rooting transferred to soil in greenhouses at the University of California-Davis at 27 °C and 50% humidity. Berries produced by open pollination were collected from greenhouse plants. Berries were also collected in the field from R. armeniacus and R. ursinus at DVS. Berries were transported to the lab where they were crushed and repeatedly washed to separate seeds from fruit pulp. Nineteen to 27 seeds per clone were genotyped directly. To obtain seedlings for genotyping, 50 seeds from each of the three hybrid clones and three parental species were scarified in 1% sodium hypochlorite for 24 h then left at room temperature for 90 days, followed by 90 days at 4 °C (Brinkman, 1974). Seeds were then germinated on moist Whatman paper in Petri dishes and leaf tissue was collected from seedlings for DNA extraction. Four seedlings were obtained from R. ursinus × R. pensilvanicus, and nine from one of the R. ursinus × R. armeniacus clones.
DNA extraction and molecular markers
DNA from field and cultivated samples and seedlings was extracted from leaves using a cetyltrimethylammonium bromide procedure modified from Bousquet et al. (1990). Additional β-mercaptoethanol (up to 0.5%) or polyvinylpyrrolidone-40 (0.02 g ml−1) was added to the extraction buffer if parts of tissue were red or brown in color. Approximately 2 cm2 of leaf tissue was ground under liquid nitrogen and added to 0.65 ml extraction buffer before incubation, chloroform extraction and precipitation as described by Bousquet et al. (1990). Seed embryo DNA was obtained by direct extraction from seeds, which in Rubus contain very little endosperm (Brinkman, 1974). Individual seeds were placed between two disposable plastic sheets and cracked open with a hammer, then extracted using the CTAB method or with a ZR Plant/Seed DNA MicroPrep (Zymo Research Corporation, Irvine, CA, USA). For all samples, DNA was quantified on a 1% agarose gel and diluted to a working concentration of 10 ng μl−1.
Microsatellite markers RUB126, RUB262, RUB26 (Graham et al., 2002), RhCBA6, RhCBA14, RhCBA15, RhCBA23 and RhCBA28 (Lopes et al., 2006) were used to genotype field and cultivated samples on an ABI 3100 Genetic Analyzer (Life Technologies Corporation, Carlsbad, CA, USA). Annealing temperatures were 54 °C for RhCBA15, 66 °C for RhCBA6 and 59 °C for all others. Genotypes were scored using Applied Biosystems GeneMapper 3.7 software (Life Technology Corporation) and manually corrected. Allele copy number was assumed to be ambiguous when a single-locus genotype had more than one allele but fewer alleles than the maximum number, given the ploidy. Seeds and seedlings used to determine reproductive mode were genotyped with RUB262, RhCBA14, RhCBA15, RhCBA23 and RhCBA28 only because of the high heterozygosity and reproducibility of these loci.
Additionally, field and cultivated samples were genotyped using two chloroplast markers. On the basis of the sequence polymorphism identified in GenBank (Potter et al., 2002), a cleaved amplified polymorphic sequence marker was developed to distinguish the R. ursinus chloroplast haplotype from all other Rubus chloroplast haplotypes. This marker was assayed by PCR amplification using the primers trnK685F and trnK2R, and thermal cycling program from Hu et al. (2000) followed by digestion with HindIII and visualization on 1.5% agarose gels. A single 1.8-kb band (‘uncut’ allele) was indicative of R. ursinus, whereas all other species gave bands at 1.1 and 0.7 kb (‘cut’ allele). The second chloroplast marker reflected polymorphism in length in the ndhF region of the chloroplast genome (Howarth et al., 1997). The primers 5′-GGCGGTCGAATTTCTTCTTA-3′ and 5′-ATCCTTCCCCTTCCCTTTTT-3′ were labeled with 6-carboxy-fluo-rescine and amplified at an annealing temperature of 54 °C in a multiplex with RhCBA15. Electrophoresis and scoring of fragment length for the ndhF locus was done alongside the microsatellite markers.
Data analyses
Detection of hybridization
Samples were divided into taxonomic groups based on morphology (Ertter, 1993) and chloroplast haplotype. Although some sites consisted of only one species, this is unlikely to be a guarantee of genetic isolation as seeds are bird-dispersed and there is a high amount of sympatry and overlap in the geographic ranges of the species (Supplementary Table 1). Therefore, distance-based clustering methods were used to identify hybrids, as opposed to the identification of species-specific alleles. Pairwise genetic distances between all samples were calculated from microsatellite data using the Bruvo.distance measure with the meandistance.matrix function in the R package POLYSAT 1.1 (Clark and Jasieniuk, 2011). This is a modified version of the method of Bruvo et al. (2004), which counts each allele that is present as being present in one copy and therefore does not require knowledge of ploidy or iteration through all possible genotypes where allele copy number is ambiguous. The result is a measure similar to band-sharing measures used with dominant data, but with mutational distances between microsatellite alleles also taken into account.
To focus the analysis on species that were potentially hybridizing, distributions of distances between and within taxonomic groups were used to determine a threshold distance for potential hybridization. Pairs of groups with a minimum between group distance below this threshold were subjected to Principal Coordinates Analysis (PCoA) using the cmdscale function in R (R Development Core Team, 2011). Scree plots were used to visualize the appropriate number of principal coordinates to use in the analysis.
Detection of introgression
To identify introgression of cultivated and hybrid material into the most widespread native species, all samples with the ‘uncut’ trnK allele, that is, all samples maternally derived from R. ursinus (Table 1), were subjected to Bayesian clustering analysis using the program STRUCTURE 2.3.3 (Hubisz et al., 2009). Although R. ursinus, an allopolyploid, violates the assumption of polysomic inheritance, the analysis still generated biologically meaningful clusters, for example, all cultivated samples consistently had high Q values in one cluster. As the dataset included many ambiguous polyploid genotypes, the recessive alleles model was used and the recessive allele symbol was set to the missing data symbol. POLYSAT (Clark and Jasieniuk, 2011) was used to estimate the ploidy of each individual based on allele counts and to generate the data file for STRUCTURE. To accommodate the ploidy estimated for most R. ursinus individuals without increasing the computation time to include uncommon dodecaploid individuals, the ploidy of the data file was set to eight. Individuals with an estimated ploidy less than eight had missing data symbols inserted to indicate the difference in ploidy, and rare genotypes with more than eight alleles had alleles removed at random by the write.Structure function. STRUCTURE runs had 400 000 MCMC (Monte Carlo Markov Chain) repetitions after a burn-in of 20 000 repetitions under default parameters. At least three runs were performed at each K value for K=1 through 8. The method of Evanno et al. (2005), in conjunction with biological meaningfulness of the results, was used to determine the correct K value.
Determining the reproductive mode of hybrids
Microsatellite data were imported into the R package POLYSAT (Clark and Jasieniuk, 2011), and analysis was performed in R to assess the degree of similarity between maternal and offspring genotypes. Given the polyploid nature of the data and the low quantity of DNA derived from seed embryos, allelic dropout was considered likely to cause some artificial differences between the genotypes of asexually related individuals at a few loci. However, offspring that were the product of sexual recombination would be expected to have a different genotype from the mother at most loci. Therefore, the dissimilarity between mother and offspring was measured as the number of loci at which their genotypes differed. R. armeniacus and R. pensilvanicus served as controls for the distribution of dissimilarities under apomixis, whereas R. ursinus served as a control for the dissimilarity distribution under sexual reproduction.
Results
Species, cultivars and microspecies aggregate sampled
The distribution of species sampled across field sites, based on morphological characters (Ertter, 1993) is summarized in Supplementary Table 1. Nearly all sites included either the native blackberry (subgenus Rubus) R. ursinus or the highly invasive blackberry R. armeniacus, with both species growing sympatrically at many sites. R. armeniacus is a microspecies within the apomictic R. fruticosus aggregate, all of which are native to Eurasia, and for the purposes of this study is grouped with several other R. fruticosus agg. microspecies that were found less frequently. One other exotic blackberry, R. pensilvanicus, was found at two sites in the Sacramento Valley. Two native raspberries (subgenus Idaeobatus), R. spectabilis and R. leucodermis, were found occasionally in the northern end of our sampling range and in the Sierra Nevada, and the native thimbleberry (subgenus Anoplobatus) R. parviflorus was found occasionally in northern coastal California. Cultivated varieties collected included raspberry (subgenus Idaeobatus) and blackberry (subgenus Rubus) varieties, as well as hybrid types (that is, Logan, Boysen, Marion, Ollalie, Tay, Siskiyou and Silvan) derived from R. ursinus, R. fruticosus agg. and other species of the subgenera Rubus and Idaeobatus.
Marker polymorphism
Two PCR-based chloroplast markers and eight nuclear microsatellite markers (Tables 1 and 2) were used to genotype all field collections. The trnK marker was very effective at distinguishing the R. ursinus chloroplast genome from that of any other species (Table 1). Within the other species, the ndhF chloroplast marker had distinct size ranges for R. fruticosus agg. (340–346 bp) and R. pensilvanicus (356 bp), whereas R. parviflorus and all species of the subgenus Idaeobatus had shared alleles (330–338 bp) (Table 1). Many alleles were detected at each of the eight nuclear microsatellite loci, particularly within the native R. ursinus (Table 2).
Evidence of hybridization
Interspecific hybrids were detected by analysis of genetic distances between individuals. On the basis of trnK and ndhF chloroplast alleles (Table 1) and morphology, we were able to assign all individuals to one of six species or a group containing all cultivated varieties. Comparisons of inter-individual distances between and within groups indicated that 0.6 was the minimum distance that would typically be observed between individuals of different species (Supplementary Figure 1). Thus, all pairs of groups with a minimum distance of less than 0.6 between individuals of different groups (Supplementary Table 2) were considered to be potentially hybridizing and were subjected to PCoA for further examination. Eigenvalues, as visualized in scree plots, declined sharply after the first two principal coordinates for all pairs (Supplementary Figure 2), with the exception of R. ursinus vs R. spectabilis and R. ursinus vs R. leucodermis, for which eigenvalues declined sharply after the first four principal coordinates, indicating that biologically meaningful results would be seen using the first two or four coordinates, respectively.
R. ursinus, R. fruticosus agg. and R. pensilvanicus were strongly differentiated by PCoA, with hybrid clusters present directly between and distinct from the parent clusters (Figures 2a and b). Both of these clusters represent hybridization between exotic and native species, and in both cases the native, sexually reproducing R. ursinus is the maternal parent, which is unsurprising given that the other parent species produce most of their seeds by apomixis. Within R. fruticosus agg., R. armeniacus is the paternal parent of the hybrids with R. ursinus based on the positions of the clusters (Figure 2a).
Principal coordinate analyses of selected Rubus groups. The first two principal coordinate axes are shown, with the percentage of variation represented by the axis given in parentheses. Individuals with the ‘cut’ trnK allele are shown in red, and individuals with the ‘uncut’ chloroplast trnK allele (characteristic of R. ursinus) are shown in blue. Symbols represent different chloroplast ndhF alleles. (a) R. ursinus vs R. fruticosus agg. (b) R. ursinus vs R. pensilvanicus. (c) R. ursinus vs cultivated varieties. Cultivated varieties with the ‘uncut’ trnK allele are marked with green circles. (d) R. leucodermis vs cultivated varieties. R. leucodermis is marked with purple circles, and cultivated raspberry varieties are marked with green circles.
PCoA also suggested some hybridization between native species and cultivated varieties. There was considerable overlap between the clusters of cultivated varieties and R. ursinus (Figure 2c), although the only cultivated varieties with a distance of less than 0.6 to R. ursinus were those maternally derived from R. ursinus (Supplementary Figure 1c). R. leucodermis formed an elongated cluster that extended toward the cluster of cultivated raspberries (Figure 4d), suggesting hybridization with backcrossing to R. leucodermis. This is consistent with R. leucodermis and cultivated raspberries both being sexual diploids in the same subgenus. PCoA of other pairs of groups did not indicate hybridization (Supplementary Figure 3).
Evidence of introgression in R. ursinus
All individuals with the ‘uncut’ trnK allele (Table 1) were subjected to clustering analysis with the software STRUCTURE, to clarify the results of PCoA (Figure 2c) that suggested introgression of R. ursinus-derived cultivated material into native R. ursinus populations. Likelihood values were variable across runs because of the polyploid nature of the dataset, but still fit a smooth curve (Figure 3a and two outliers with lower likelihoods, not shown). When outliers were excluded, analysis of likelihoods using the method of Evanno et al. (2005) indicated that two populations should be assumed (Figure 3b). In every run, all cultivated varieties consistently had high Q values in a single cluster, allowing us to make inferences about introgression of cultivated material into R. ursinus. Results at K=2 suggested a large amount of introgression (Q=0.35) of cultivated Rubus into R. ursinus (Figure 3c), particularly in the northern area of the sampling range (Supplementary Figure 4a). However, the apparent amount of introgression was reduced significantly at larger values of K (Figure 3c, Supplementary Figures 4b and c), stabilizing at Q=0.01 to 0.02 at K=6 through 8. Because ΔK had secondary peaks at K=4 and K=6 (Figure 3b), bar plots at these K values are shown in Supplementary Figures 4b and c to illustrate the decrease in apparent introgression. Individual Q values for all runs shown in Figure 3 are provided in Supplementary Material S2. Approximately 2% of R. ursinus individuals consistently showed signs of introgression from cultivated material in all runs despite multimodality at K values greater than 2 (Figure 3e). These were found at sites KVY, ARC and ARP (Supplementary Material S2). In the nine runs that placed R. ursinus × R. pensilvanicus hybrids into a separate cluster (Supplementary Figure 4c, Supplementary Material S2), 3% of R. ursinus individuals consistently showed signs of introgression from R. pensilvanicus (Figures 3d and f), including individuals at sites BWP, KNL, ARP, MCD and CTF (Supplementary Material S2). R. ursinus × R. armeniacus individuals were consistently placed in a cluster with wild R. ursinus (Supplementary Figure 4), so evidence of introgression of R. armeniacus into R. ursinus could not be assessed.
Summary of STRUCTURE results for K=1 through 8. (a) Expected log probability of the data for all runs, excluding two outliers with lower values. (b) ΔK as determined with the method of Evanno et al. (2005). (c) Mean Q values of the cluster corresponding to cultivated Rubus, within wild R. ursinus excluding R. ursinus × R. armeniacus and R. ursinus × R. pensilvanicus hybrids. Lines indicate the standard deviation of this Q value within each run. Mean values are interpreted as the estimated amount of introgression of cultivated Rubus into wild R. ursinus. (d) Mean Q values of the cluster corresponding to R. ursinus × R. pensilvanicus, within wild R. ursinus as in part (c). Only the nine runs in which R. ursinus × R. pensilvanicus formed a cluster separate from cultivated Rubus are represented. (e, f) Frequencies of wild R. ursinus individuals with Q>0.1 in the cultivated or R. ursinus × R. pensilvanicus clusters, respectively. Dark gray bars indicate the frequency of individuals with Q>0.1 in at least one run at a given K value. Medium gray bars indicate the frequency of individuals with Q>0.1 in all runs at a given K value. Light gray bars indicate the frequency of individuals with Q>0.1 in all runs at a given K value and all lower K values.
Reproductive mode of hybrids
To distinguish offspring produced sexually from those produced apomictically, the number of loci was counted at which the genotype of each offspring differed from that of its mother. As expected, nearly all offspring of the sexual, diecious R. ursinus differed from the mother plant at all loci for which the mother was heterozygous (four out of five loci; Figure 4a). All other mother plants in the study were heterozygous at all five loci (data not shown). Most offspring of the apomictic R. armeniacus differed from the mother plant at zero or one loci (Figure 4b), whereas most offspring of R. pensilvanicus differed from the mother plant at two or three loci (Figure 4f).
Distributions of dissimilarities between the genotype of each progeny and its mother, which were used to determine the reproductive mode of interspecific hybrids. X-axes indicate the number of loci at which the offspring had a different genotype from the parent. Y-axes indicate the number of progeny with each number of differing loci. Shown are the results for (a) R. ursinus seeds, (b) R. armeniacus seeds, (c) R. ursinus × R. armeniacus clone CSP6A seeds, (d) R. ursinus × R. armeniacus clone CSP6A seedlings, (e) R. ursinus × R. armeniacus clone CSP2A seeds, (f) R. pensilvanicus seeds, (g) R. ursinus × R. pensilvanicus seeds and (h) R. ursinus × R. pensilvanicus seedlings.
Most seedling offspring of both the R. ursinus × R. armeniacus (Figure 4d) and R. ursinus × R. pensilvanicus (Figure 4h) differed from the mother plant at most loci, indicating predominantly sexual reproduction in these hybrids. However, when offspring of the hybrids were examined at the seed embryo stage, they differed from the mother plants at fewer loci (Figures 4c), suggesting that many inviable seeds are produced asexually.
Discussion
F1 hybrids between native and exotic Rubus
F1 R. ursinus × R. armeniacus and R. ursinus × R. pensilvanicus hybrids, the latter being unknown before our study, were unambiguously identified by PCoA. Although R. armeniacus and R. ursinus are very commonly found growing in the same sites, hybridization between them is rare based on our results. Our results may in fact overestimate the rate of hybridization, as we only sampled the one site with R. ursinus × R. armeniacus hybrids because it was brought to our attention by park managers. On the other hand, R. pensilvanicus is uncommon as an exotic species on the West Coast but, based on our results, hybridizes very readily with R. ursinus. We only found R. pensilvanicus at two of our field sites, but we also identified two field sites with R. ursinus × R. pensilvanicus hybrids (Supplementary Table 1). These results support our hypothesis by demonstrating that hybridization is occurring between native and introduced Rubus species.
R. ursinus × R. armeniacus and R. ursinus × R. pensilvanicus hybrids can be identified in the field by their morphological characteristics. Both hybrid types have leaf, stem and prickle morphology that is intermediate to that of the parents (data not shown). R. ursinus × R. pensilvanicus more closely resembles R. ursinus at first glance, but has slightly angular stems and some leaves that are more heavily dissected than those of R. ursinus. Given that all Rubus native to the West Coast has round stems, the angular stem may be used as a reliable indicator of exotic species and their hybrids, should control be desirable. Both hybrid types have hermaphroditic flowers such as R. armeniacus and R. pensilvanicus, whereas R. ursinus is diecious. Hermaphroditic flowers would be advantageous for providing mating system flexibility during the range expansion of an exotic species, as diecious plants cannot reproduce unless individuals of both sexes are present and have a reduced effective population size if the sex ratio is uneven in a small founding population.
Although these F1 hybrids are not sufficiently widespread (Supplementary Table 1) to be recognized as invasive, they could represent an early stage in the evolution of new invasive Rubus taxa. Both hybrid types grew very vigorously in the greenhouse and appear to outcompete R. ursinus at field sites. As these hybrids combine the prickle stiffness of R. armeniacus and R. pensilvanicus with the high prickle density of R. ursinus, they may be especially difficult to remove manually, although this is only one component of invasion success in Rubus. It has also been demonstrated that R. ursinus and R. armeniacus differ in their allocation of resources to vegetative and reproductive growth in ways that may affect invasiveness (Lambrecht-McDowell and Radosevich, 2005), and it is currently unknown whether the hybrids resemble the invasive parent in this respect. Indeed, R. ursinus × R. armeniacus hybrids have been observed in California for over half a century and could potentially have been in existence since the introduction of R. armeniacus in the late nineteenth century (Mallah, 1954), but have not yet become invasive, particularly when compared with the scale of the R. armeniacus invasion. However, the generation of F1 hybrids is expected to be a slow process in the case of R. ursinus × R. armeniacus, given that the flowering times of R. armeniacus and R. ursinus only overlap for a short period in California, and further north do not overlap at all (Susan Lambrecht, personal communication). Further, the possible occurrence of multiple hybrid cytotypes differing in ploidy level, which we could not assess but can result from the uncoupling of apomeiosis and parthenogenesis in facultative apomictic plants, may also be causing failed interploidal crosses and hybrid instability (Cosendai and Hörandl, 2010), impairing novel hybrids from spreading through sexual reproduction.
R. ursinus may hybridize more readily with R. pensilvanicus than R. armeniacus, but because of the rarity of R. pensilvanicus in the Western United States, hybrids are still uncommon. We expect that more invasive hybrid genotypes could be produced than those found in our study and those that have existed historically, given the rarity of hybridization and the high genetic diversity in R. ursinus. Although few clones of the apomictic species R. fruticosus agg. and R. pensilvanicus exist in our study range, these were heterozygous at most loci tested, further contributing to the diversity of hybrid genotypes that could be produced. As these hybrids are neoallopolyploids, additional evolution may occur through genomic rearrangements and epigenetic remodeling (Comai, 2005). Lag periods often exist between introduction and invasion, and it has been hypothesized that evolutionary changes occur during these lag times (Schierenbeck and Ellstrand, 2009).
Introgression of cultivated and exotic Rubus into native Rubus
Our PCoA and STRUCTURE results also suggest some hybridization of cultivated Rubus with native Rubus, as well as introgression of R. pensilvanicus into R. ursinus. Cultivated varieties derived from R. ursinus overlapped with R. ursinus in PCoA. In Bayesian clustering analysis, these cultivated varieties were placed almost entirely into one cluster, and 2% of wild R. ursinus individuals consistently had partial membership in that cluster. Additionally, PCoA suggests hybridization of the native raspberry R. leucodermis with cultivated raspberries, with backcrossing to R. leucodermis. Backcrossing would not be prevented by ploidy differences, in this case, because nearly all raspberries are diploid. PCoA did not suggest backcrossing of R. ursinus × R. armeniacus or R. ursinus × R. pensilvanicus into either parent species, although our STRUCTURE results suggest that R. ursinus × R. pensilvanicus may have backcrossed to R. ursinus to a small degree.
The amount of introgression into R. leucodermis is large enough that it may have fitness consequences for the species as a whole, whereas the minor amount of introgression into R. ursinus is of little concern. Two possible ecological consequences of such introgression are outbreeding depression and the generation of weedier genotypes through increased genetic diversity (Rhymer and Simberloff, 1996).
Reproductive mode of F1 hybrids
On the basis of genotypes of progeny of R. ursinus × R. armeniacus and R. ursinus × R. pensilvanicus F1 hybrids, these neoallopolyploids have the ability to reproduce sexually, allowing for further evolution and selection, or by apomixis, preserving highly fit heterozygous genotypes. Mallah (1954) proposed, based on observations of meiosis, that R. ursinus × R. armeniacus hybrids have three disomic homeologous genomes, allowing the hybrids to reproduce sexually without any genome doubling. When the R. ursinus × R. armeniacus hybrids of Mallah were used as the maternal parent in a cross with R. armeniacus, a few seedlings were produced that varied in morphology and vigor, also suggesting sexual recombination. Although ploidy and inheritance mode are unknown in the R. ursinus × R. armeniacus hybrids that we identified, our molecular results also indicate that their seedlings were produced by sexual recombination. Given the tendency of apomixis to be dominant and associated with polyploidy (Comai, 2005; Ozias-Akins and van Dik, 2007), it is not surprising that the F1 hybrids also retained the ability to produce some seeds apomictically. However, reversion to sexuality has been observed in hybrids between apomictic Rubus species (Nybom, 1988), suggesting complex inheritance of apomixis. The intermediate amount of apomixis observed in the hybrids in this study suggests incomplete dominance at one or more loci controlling apomixis. Loci controlling apomixis have not been mapped in the Rosaceae, but apomixis in other taxa is generally found to be under the control of multiple genes, with unlinked modifier loci affecting penetrance (Ozias-Akins and van Dik, 2007). Another possibility in our system is that the proportion of progeny produced by apomixis is an additive trait that depends on dosage of alleles: given the higher ploidy of the sexual parent, this parent is likely to have contributed more than the apomictic parent to the genomic composition of the hybrids, which could account for the predominantly sexual reproductive mode of the hybrids.
The mixture of reproductive modes gives the hybrids immediate access to multiple evolutionary options: asexual preservation of a highly fit genotype, sexual recombination by self-pollination or mating with another hybrid of the same allopolyploid genome constitution, or hybridization to create a different allopolyploid type. In addition to potential changes in fitness across generations, the rate of apomixis itself may evolve, given the large-scale genomic rearrangements and epigenetic changes that can happen early in the evolution of an allopolyploid, as well as Mendelian segregation at the loci controlling apomixis. One possible scenario is an increase in fitness as a result of sexual recombination, followed by maintenance of highly fit genotypes by an increased rate of apomixis.
The system that we characterize in this paper is similar to other instances in the invasion biology literature of hybridization followed by asexual seed production, although unique in that it involves hybridization between sexual and apomictic species. Hybrid R. alceifolius, described in the introduction of this paper, was produced by crosses between two sexual species, and appears to have undergone little to no sexual recombination in its invaded range (Amsellem et al., 2001). Amelanchier erecta, also in the Rosaceae, is a facultative apomictic hybrid species with a broader range than that of its parent species, which are also facultatively apomictic (Campbell et al., 1997). In both of these cases as well as ours, multiple hybrid clones exist in nature, indicating multiple hybridization events and/or sexual recombination post-hybridization. Studies such as ours that examine the early stages of evolution of novel hybrids shed light on how invasiveness may evolve.
Conclusion
Hybridization, followed by either asexual or sexual reproduction, can make significant contributions to the evolution of invasiveness. The diverse genus Rubus, which contains many apomictic species and is prone to invasiveness, can provide a useful system to study this phenomenon. Our study identified a small number of F1 hybrids between a native species of Rubus and two exotic species, and supported our hypothesis that these hybrids are able to reproduce by apomixis, although the majority of germinated seedlings showed evidence of sexual recombination. We also detected introgression from the gene pool of cultivated Rubus into multiple native species. This adds to a growing body of research on the evolutionary consequences of anthropogenic movement of species, including biological invasions and changes to the genetic composition of native species (Suaraz and Tsutsui, 2008). Future studies of reproductive mode changes caused by hybridization events in other taxa will yield insight into the importance of such occurrences in the evolution of invasiveness.
Data archiving
Data have been deposited at Dryad: doi:10.5061/dryad.m466f.
References
Amsellem L, Noyer JL, Hossaert-McKey M (2001). Evidence for a switch in the reproductive biology of Rubus alceifolius (Rosaceae) towards apomixis, between its native range and its area of introduction. Am J Bot 88: 2243–2251.
Baker HG (1965). Characteristics and modes of origin of weeds. In: Baker HG, Stebbins LG (eds) The Genetics of Colonizing Species. Academic Press: New York, pp 147–172.
Barrett SCH, Colautti RI, Eckert CG (2008). Plant reproductive systems and evolution during biological invasion. Mol Ecol 17: 373–383.
Bousquet L, Simon L, Lalonde M (1990). DNA amplification from vegetative and sexual tissues of trees using polymerase chain reaction. Can J For Res 20: 254–257.
Brinkman KA (1974). Rubus L. In: Schopmeyer CS (ed.) Seeds of Woody Plants in the United States. U.S. Department of Agriculture, Forest Service: Washington, DC, pp 738–743.
Brown SW (1943). The origin and nature of variability in Pacific coast blackberries (Rubus ursinus Cham & Schlecht and R. lemurum Sp Nov). Am J Bot 30: 686–697.
Bruvo R, Michiels NK, D’Souza TG, Schulenburg H (2004). A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13: 2101–2016.
Campbell CS, Wojciechowski MF, Baldwin BG, Alice LA, Donoghue MJ (1997). Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex (Rosaceae). Mol Biol Evol 14: 81–90.
Chase MW, Hills HH (1991). Silica-gel—an ideal material for field preservation of leaf samples for DNA studies. Taxon 40: 215–220.
Clark LV, Jasieniuk M (2011). POLYSAT: an R package for polyploid microsatellite analysis. Mol Ecol Resour 11: 562–566.
Comai L (2005). The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846.
Cosendai AC, Hörandl E (2010). Cytotype stability, facultative apomixes and geographical parthenogenesis in Ranunculus kuepferi (Ranunculaceae). Ann Bot 105: 457–470.
Crane MB (1940). Reproductive versatility in Rubus I. Morphology and inheritance. J Genet 40: 109–118.
Ertter B (1993). Rubus. In: Hickman JC (ed) The Jepson Manual: Higher Plants of California. University of California Press: Berkeley, pp 974–975.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620.
Evans KJ, Weber HE (2003). Rubus anglocandicans (Rosaceae) is the most widespread taxon of European blackberry in Australia. Aust Syst Bot 16: 527–537.
Geng YP, Pan XY, Xu CY, Zhang WJ, Li B, Chen JK et al (2007). Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol Invasions 9: 245–256.
Graham J, Smith K, Woodhead M, Russell J (2002). Development and use of simple sequence repeat markers in Rubus species. Mol Ecol Notes 2: 250–252.
Howarth DG, Gardner DE, Morden CW (1997). Phylogeny of Rubus subgenus Idaeobatus (Rosaceae) and its implications toward colonization of the Hawaiian Islands. Syst Bot 22: 433–441.
Hu JM, Lavin M, Wojciechowski MF, Sanderson MJ (2000). Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on chloroplast trnK/matK sequences and its implications for evolutionary patterns in Papilionoideae. Am J Bot 87: 418–430.
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009). Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9: 1322–1332.
Lambrecht-McDowell SC, Radosevich SR (2005). Population demographics and trade-offs to reproduction of an invasive and noninvasive species of Rubus. Biol Invasions 7: 281–295.
Lee CE (2002). Evolutionary genetics of invasive species. Trends Ecol Evol 17: 386–391.
Lopes MS, Maciel G, Mendonca D, Gil FS, Machado ADC (2006). Isolation and characterization of simple sequence repeat loci in Rubus hochstetterorum and their use in other species from the Rosaceae family. Mol Ecol Notes 6: 750–752.
Mallah TS (1954) Natural hybridization between Rubus ursinus Cham & Schlecht and other Rubi in California. Ph.D. Dissertation, University of California.
McGregor G (1998). Relationships between weedy and commercially grown Rubus species. Plant Prot Q 13: 157–159.
Novak SJ, Mack RN (2005). Genetic bottlenecks in alien plant species: Influence of mating systems and introduction dynamics. In: Sax DF, Stachowicz JJ, Gaines SD (eds) Species Invasions: Insights into Ecology, Evolution, and Biogeography. Sinauer Associates: Sunderland, MA, pp 201–228.
Nybom H (1985). Active self-pollination in blackberries (Rubus subgen Rubus, Rosaceae). Nord J Bot 5: 521–525.
Nybom H (1988). Apomixis vs sexuality in blackberries (Rubus subgen Rubus, Rosaceae). Plant Syst Evol 160: 207–218.
Nybom H, Schaal BA (1990). DNA fingerprints reveal genotypic distribution in natural populations of blackberries and raspberries (Rubus, Rosaceae). Am J Bot 77: 883–888.
Ozias-Akins P, van Dik PJ (2007). Mendelian genetics of apomixis in plants. Annu Rev Genet 41: 509–537.
Potter D, Gao F, Bortiri PE, Oh SH, Baggett S (2002). Phylogenetic relationships in Rosaceae inferred from chloroplast matK and trnL-trnF nucleotide sequence data. Plant Syst Evol 231: 77–89.
Poulin J, Weller SG, Sakai AK (2005). Genetic diversity does not affect the invasiveness of fountain grass (Pennisetum setaceum) in Arizona, California and Hawaii. Divers Distrib 11: 241–247.
R Development Core Team (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
Rhymer JM, Simberloff D (1996). Extinction by hybridization and introgression. Ann Rev Ecol Syst 27: 83–109.
Schierenbeck KA, Ellstrand NC (2009). Hybridization and the evolution of invasiveness in plants and other organisms. Biol Invasions 11: 1093–1105.
Shannon CE (1948). A mathematical theory of communication. Bell Syst Technical J 27: 379–423.
Springer NM, Stupar RM (2007). Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Res 17: 264–275.
Suaraz AV, Tsutsui ND (2008). The evolutionary consequences of biological invasions. Mol Ecol 17: 351–360.
Thompson MM (1995). Chromosome numbers of Rubus species at the National Clonal Germplasm Repository. HortScience 30: 1447–1452.
United States Department of Agriculture (2011). PLANTS Database. National Plant Data Center, Baton Rouge: Louisiana.
Weber HE (1996). Former and modern taxonomic treatment of the apomictic Rubus complex. Folia Geobot Phytotaxon 31: 373–380.
Acknowledgements
We thank Stella Hartono for assistance in the lab and field. Bernie May, Kristina Schierenbeck, Jeff Firestone, Miki Okada, the subject editor and two anonymous reviewers provided feedback on an earlier version of this manuscript. Kristina Schierenbeck, Lawrence Alice and Marcel Rejmanek made recommendations and provided assistance in the early stages of this study. Susan Lambrecht, Ramona Robison, Jeff Firestone, Anna Sherwood, Annabelle Kleist, Noor-ul-ain Noor, Cosumnes River Preserve, Caswell Memorial State Park, American River Cherry Company, Pacific Star Gardens, Brooklyn Botanic Garden, Coal Oil Point Reserve, Paradise Reserve and Sedgwick Reserve gave assistance and/or permissions for field collections. Research funds were provided by a Jastro-Shields Research Award and a California Weed Science Society scholarship. LVC was supported by a UC Davis Department of Plant Sciences Research Assistantship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Clark, L., Jasieniuk, M. Spontaneous hybrids between native and exotic Rubus in the Western United States produce offspring both by apomixis and by sexual recombination. Heredity 109, 320–328 (2012). https://doi.org/10.1038/hdy.2012.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.45
Keywords
This article is cited by
-
SSR fingerprinting of a German Rubus collection and pedigree based evaluation on trueness-to-type
Genetic Resources and Crop Evolution (2017)
-
Analysis of Allelic Diversity and Genetic Relationships Among Cultivated Mangosteen (Garcinia mangostana L.) in Java, Indonesia Using Microsatellite Markers and Morphological Characters
Tropical Plant Biology (2016)